Neurodegenerative diseases are among the most pressing and devastating health challenges facing aging populations globally. Stem-cell technologies now enable us to generate brain tissue in the lab from our patients with these diseases. We utilize these patient-specific models to better understand the origin and consequences of disease processes, and to develop personalized treatments.
The Khurana Lab is located within the Ann Romney Center for Neurologic Diseases at Brigham and Women’s Hospital and Harvard Medical School. Together with our local, national and international collaborators, we strive to better understand and develop therapies for some of the most detrimental age-related disorders of our time: neurodegenerative diseases.
CENTRAL PROBLEM
Understandably, each patient diagnosed with a neurodegenerative disease has a burning question: “Why me?” We believe every patient deserves the answer to that question. Neurodegenerative diseases are heterogeneous. We believe that the same disease, for example Parkinson’s disease, can be triggered in different ways in different patients. This means that a “one size fits all strategy” may not work for therapy. Our lab aims to develop methods to subtype our patients, to classify each one according to the specific trigger and answer that “Why me?” question. This understanding will ultimately enable us to develop personalized medicines: we will be able to appropriately match a patient to the right drug.
UNIQUE PATIENT POPULATIONS
Thanks to the generosity of patients and their caregivers, we generate our genomes, biomarkers, imaging and stem-cell data from patients that we know and care for. This allows us to ask questions related to the specific patient being investigated, and enables our personalized medicine approach. In some cases, we search far and wide for the right patients to study. Patients with multiple system atrophy (MSA) visit us nationally and internationally. We partner directly with patients harboring rare alpha-synuclein mutations. For example, we closely work with a Spanish kindred in Northern Spain harboring the alpha-synuclein E46K mutation. For polyglutamine expansion disorders, we are developing gene therapies for specific patients with our collaborators and testing them in their matched stem-cell models.
DISEASE FOCUS
The approaches we develop are readily applicable to all neurodegenerative diseases, including Alzheimer’s and Parkinson’s disease. Our current projects focus on:
- synucleinopathies like Parkinson’s disease, multiple system atrophy and dementia with Lewy bodies
- cerebellar ataxias, from sporadic ataxias like multiple system atrophy, to hereditary ataxias like spinocerebellar ataxias, especially spinocerebellar ataxia type 3 (SCA-3) and dentatorubropallidoluysian atrophy (DRPLA).
For more specific details, including our funding sources, read below.
While neurodegeneration may originate differently in different patients, there is one common denominator: each disease is defined by the abnormal accumulation or mislocalization of a particular protein. In Parkinson’s disease, for example, that protein is called alpha-synuclein. We focus on this “common denominator” of protein misfolding in the multitude of approaches we use:
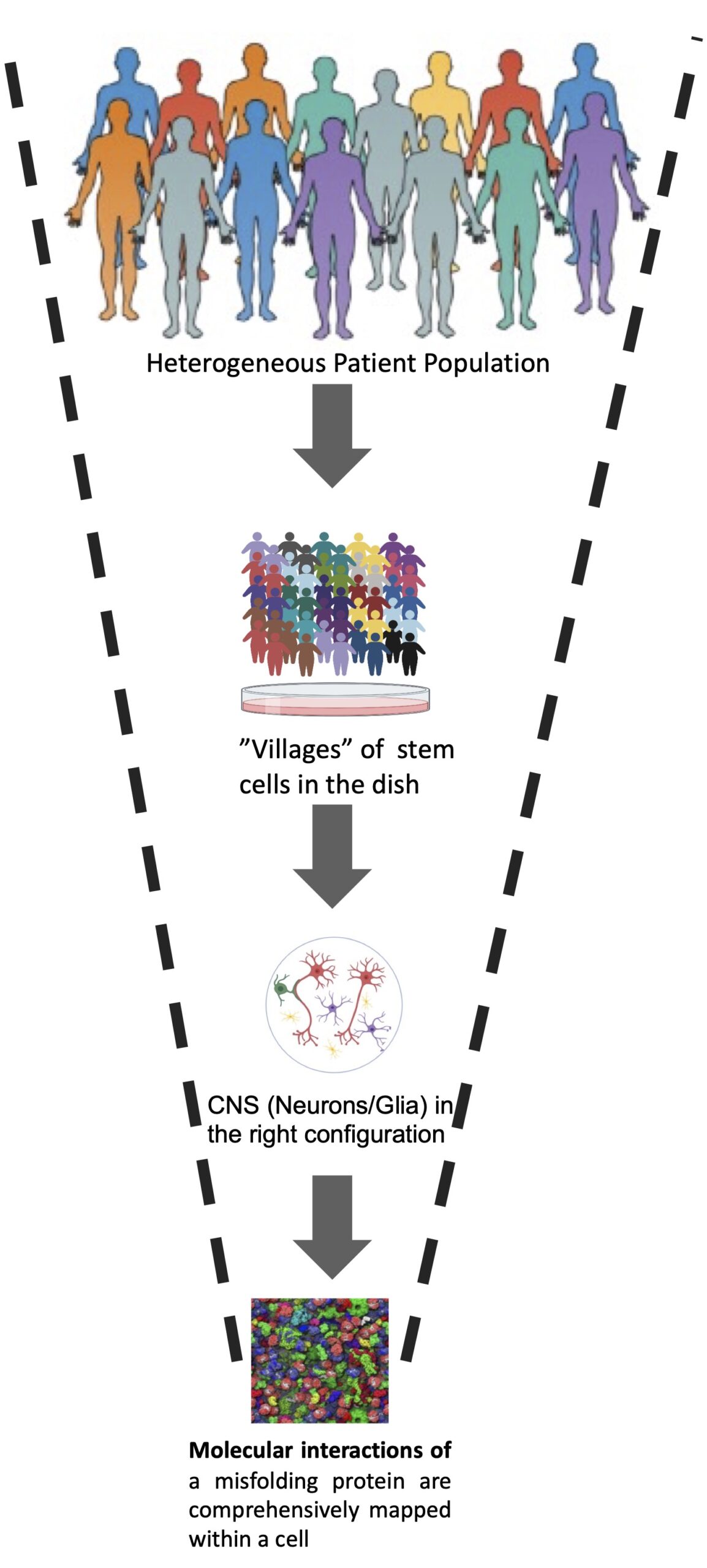
“SYSTEMS CELL BIOLOGY” APPROACH: In induced pluripotent stem cells (iPSc) from patients with neurodegenerative disease we catalog all of the key interactions (genetic and physical) of these aggregation-prone proteins in different cells of the nervous system (Fig. 1). We use these “maps” to read our patient’s genomes for clues as to the origin of the disease in that patient. We factor in their environmental exposures, from toxicants in pesticides to microbes in their gut. Our Publications page lists our recent studies. Specific methods include:
FIG. 1 The concept of genetic and spatial mapping of proteotoxicity (adapted from Jarosz and Khurana Cell 2017). A genetic map is a molecular network encompassing genes that impact a proteotoxicity when overexpressed or deleted. A spatial map comprises proteins that are in the immediate vicinity of a protein of interest. A schematic diagram of an integrated network is shown at right. Recently, such maps were generated for a-syn proteoxicity (a genetic map in yeast and a spatial map in neurons; Khurana et al., 2017; Chung et al., 2017), revealing a connection between this toxicity and 12 known parkinsonism genes. The significant overlap of genetic and spatial maps revealed an intimate relationship of a-syn toxicity to its functional interactions and location.
FIG. 2 Glio-neuronal co-cultures from entire populations of patients will ultimately be captured in a single “village”, ushering in possibilities of clinical trials “in the dish. Molecular-level knowledge of protein misfoylding will guide identification and reversal of pathologic phenotypes in these villages.